Coding Phase 3.1
Hello Everyone!
This week I worked further on the implementation of XitoSBML-CUI. I had been able to use the picocli library to get the file path of the image to be converted into an SBML document and obtain an ImagePlus object from it last week. So, now it became important to take the file path as input from the command line and obtain an SBML document as output without using any GUI functionality. It was already decided that since all the images in the dataset created for this project were segmented only for the cytosol, we would only need "Cytosol" as the default domain type in XitoSBML-CUI.
To start with I decided to write a new main class which would create an ImagePlus object and then would pass it as a parameter for getting the SBML document instead of requiring an Image Table to obtain ImagePlus object. For example, it could be done like this:
public class TrialRunner {
public static void runs(ImagePlus imager) {
// imager.show();
new MainImgSpatial().runner(imager);
}
public static void main(String[] args) {
String path = "C:\\Users\\Subroto\\Desktop\\Inference(300x100)\\gt_5.tif";
// new ij.ImageJ();
ImagePlus imager = new ImagePlus(path);
// img.show();
String title = imager.getTitle();
System.out.println(title);
runs(imager);
}
}
The current implementation of the main method is, however, CuiRun.java which takes input
from the command line instead of hard-coding it.
public class CuiRun implements Callable<Integer> {
@Option(names = "-i", required = true, description = "The path to input image file")
String inputValue;
@Option(names = "-o", required = true, description = "The path to output XML file")
String outputValue;
@Override public Integer call() {
// System.out.printf("-i=%s%n", inputValue);
// System.out.printf("-o=%s%n", outputValue);
new CuiMainImgSpatial().runCui(inputValue, outputValue);
return 0;
}
public static void main(String... args) {
System.exit(new CommandLine(new CuiRun()).execute(args));
}
}
Reference: https://github.com/Medha-B/XitoSBML/blob/xitosbml_cui/src/main/java/jp/ac/keio/bio/fun/xitosbml/cui/CuiRun.java
In the original XitoSBML code, the main class is Spatial_Img_SBML.java, which launches ImageJ and calls "run Spatial Image SBML plugin" via MainImgSpatial.java. MainImgSpatial.java extends MainSpatial.java and implements the "run Spatial Image SBML plugin" function. In the MainSpatial.java class, the gui() function gets called which implements the ImageExplorer.java class. It is through this class that the ImagePlus object (i.e. the segmented image) is taken as an input and stored along with the selected domain in a hashmap called the hashDomFile.
So, in order to change XitoSBML to XitoSBML-CUI, the first step was to put "Cytosol" as the default domain type in the hashDomFile along with the ImagePlus object. This is implemented via function getDomFile() and actionPerf() in the class TrialForImag.java. Please refer to https://github.com/Medha-B/XitoSBML/blob/xitosbml_cui/src/main/java/jp/ac/keio/bio/fun/xitosbml/cui/TrialForImg.java.
At the moment, there are four classes in the new package jp.ac.keio.bio.fun.xitosbml.cui: CuiRun.java, CuiMainSpatial.java, CuiMainImgSpatial.java and TrialForImage.java. For reference: https://github.com/Medha-B/XitoSBML/tree/xitosbml_cui/src/main/java/jp/ac/keio/bio/fun/xitosbml/cui.
Although I have not yet run CuiRun.java as a Runnable Jar file from the command line, I used the "Program Arguments" window in "Run Configurations" of Eclipse to provide the arguments. |
| Resulting SBML document gets printed |
I run the code with a few example images. These images, the resulting SBML documents, and spatial images can be obtained at https://drive.google.com/drive/folders/1-uX5HFjyW-e5-NnNUowwr5LiOEVh91YE?usp=sharing (https://drive.google.com/drive/folders/1-uX5HFjyW-e5-NnNUowwr5LiOEVh91YE)
The initial pull request https://github.com/Medha-B/XitoSBML/pull/1 brought a few issues to light. These issues are:
https://github.com/Medha-B/XitoSBML/issues/2
https://github.com/Medha-B/XitoSBML/issues/3
https://github.com/Medha-B/XitoSBML/issues/4
https://github.com/Medha-B/XitoSBML/issues/5
https://github.com/Medha-B/XitoSBML/issues/6
https://github.com/Medha-B/XitoSBML/issues/8
So, I made a new pull request to address these issues: https://github.com/Medha-B/XitoSBML/pull/7
I also worked on the issue https://github.com/Medha-B/unetdl4j/issues/16 for Inference.java in https://github.com/Medha-B/unetdl4j. Please refer to the commit https://github.com/Medha-B/unetdl4j/commit/9dab1a44f9a8bda6782cdf344243e63ed073f96a to see the changes.
Until the next time...
Alveda!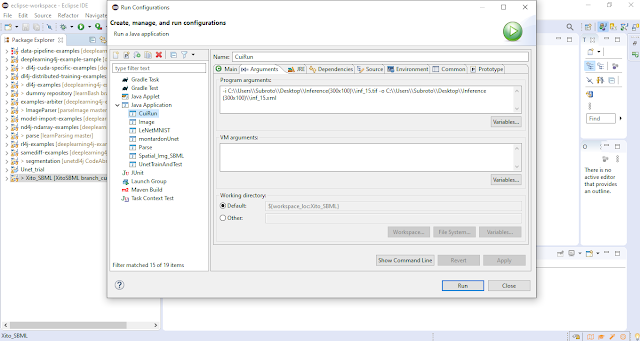

Comments
Post a Comment