Coding Phase 2.2
Hello everyone!
This week I tried to develop a method for cross-validation of the U-Net model. While a method based on Intersection-Over-Union and Dice Coefficient was developed last week for overall model evaluation, it is important to get an assurance of the accuracy of the model predictions. For this, we need to validate our model by testing it on unseen data and the results tell us whether the model is well generalized or not.
An easy way to do this is k-fold cross-validation, a technique commonly used in machine learning that estimates the skill of a model on new data. As a general procedure, the input dataset gets split into k folds, (k-1) folds get trained at a time and the remaining fold is used for validation. This process is iterated for k times and the model with the best accuracy gets selected.
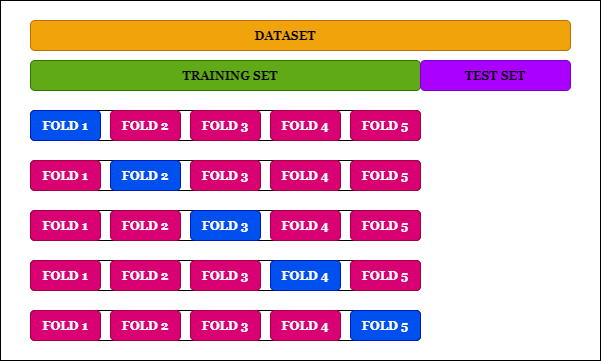
The distinctive feature of k-fold cross-validation as mentioned above is splitting of the input dataset into k parts. Although this can be done manually, we can do it using code as well.
The entire procedure for shuffling the input dataset and then splitting it into k groups can be done as follows:
File rootDir = new File("C:\\Users\\dataset");
double[] folds = new double[k];
The complete code can be obtained at https://github.com/Medha-B/unetdl4j/blob/master/src/main/java/org/sbml/spatial/segmentation/crossVal.java (commit f014e22 ).
This week I tried to develop a method for cross-validation of the U-Net model. While a method based on Intersection-Over-Union and Dice Coefficient was developed last week for overall model evaluation, it is important to get an assurance of the accuracy of the model predictions. For this, we need to validate our model by testing it on unseen data and the results tell us whether the model is well generalized or not.
An easy way to do this is k-fold cross-validation, a technique commonly used in machine learning that estimates the skill of a model on new data. As a general procedure, the input dataset gets split into k folds, (k-1) folds get trained at a time and the remaining fold is used for validation. This process is iterated for k times and the model with the best accuracy gets selected.

The distinctive feature of k-fold cross-validation as mentioned above is splitting of the input dataset into k parts. Although this can be done manually, we can do it using code as well.
The entire procedure for shuffling the input dataset and then splitting it into k groups can be done as follows:
File rootDir = new File("C:\\Users\\dataset");
FileSplit fileSplit = new FileSplit(rootDir,allowedExtensions,rng); BalancedPathFilter pathFilter = new BalancedPathFilter(rng, allowedExtensions, labeler);
For splitting the input file
For splitting the input file
double[] folds = new double[k];
Arrays.fill(folds, size_dataset / k);
InputSplit[] filesInDirSplit = fileSplit.sample(pathFilter, folds);
ImageRecordReader[] imageReader = new ImageRecordReader[k];
ImageRecordReader[] imageReader = new ImageRecordReader[k];
for( i = 0; i < k; i++) {
imageReader[i] = new ImageRecordReader(HEIGHT,WIDTH,CHANNELS,labeler);
try {
try {
imageReader[i].initialize(filesInDirSplit[i]);
}
catch (IOException e) {
catch (IOException e) {
e.printStackTrace();
}
}
RecordReaderDataSetIterator[] set = new RecordReaderDataSetIterator[k];
}
RecordReaderDataSetIterator[] set = new RecordReaderDataSetIterator[k];
for( i = 0; i < k; i++){
set[i] = new RecordReaderDataSetIterator(imageReader[i], batchSize, 1, 1, true);
scaler.fit(set[i]);
set[i].setPreProcessor(scaler);
}
At the time of training, one of the k sets will be held-out while the rest of the sets will be trained together. This process will iterate k times, by holding-out each set exactly once. The code for training can be written as
At the time of training, one of the k sets will be held-out while the rest of the sets will be trained together. This process will iterate k times, by holding-out each set exactly once. The code for training can be written as
while(testFold<k){
model.reset();
for(i=0; i<k; i++) {
if(i==testFold){
continue; //Model is not trained on the testfold
}
else
{
model.addListeners(new ScoreIterationListener());
model.fit(set[i], numEpochs);
}
}
}
Similarly, for saving the model
File locationTosave = new File("C:\\Users\\unetSave"+ "["+ testFold +"]" + ".zip"); //So that I know which testFold to test this model against
boolean saveUpdater = false;
ModelSerializer.writeModel(model,locationTosave,saveUpdater);
model.reset();
for(i=0; i<k; i++) {
if(i==testFold){
continue; //Model is not trained on the testfold
}
else
{
model.addListeners(new ScoreIterationListener());
model.fit(set[i], numEpochs);
}
}
}
Similarly, for saving the model
File locationTosave = new File("C:\\Users\\unetSave"+ "["+ testFold +"]" + ".zip"); //So that I know which testFold to test this model against
boolean saveUpdater = false;
ModelSerializer.writeModel(model,locationTosave,saveUpdater);
The complete code can be obtained at https://github.com/Medha-B/unetdl4j/blob/master/src/main/java/org/sbml/spatial/segmentation/crossVal.java (commit f014e22 ).
Testing for U-Net is equivalent to evaluation by finding the average IOU and Dice Coefficient of inferred images and ground truth images. So, for validation, we will have to use the cell images from the validation fold for inference and then get IOU and Dice Coefficient. This is slightly more complex than training or model saving and thus, will require a bit more time.
As a precursor to using the code with a larger dataset (500 training images (400 train, 100 validation) and 200 testing images), I used 44 cell images and corresponding ground truth images (4-fold cross-validation). For this, I split the dataset into 4 parts manually and shuffled the content. The training sets and corresponding held-out sets can be obtained at https://drive.google.com/drive/folders/1eyRLg1s110ID-T8Oa4Je0xB9fqyy-zZr?usp=sharing
I am currently working on writing a function to give mean IOU value for the held-out set per model for its evaluation.
It is important to note here that although it might seem like a good idea to use the dl4j API KFoldIterator to split the dataset, it does not work out in this case. Using the KFoldIterator an argument to model.fit() gives an error. This might be due to the difference in the configuration of data within this iterator, which may not be suitable for a model structure composed of CNNs.
The IOU and Dice Coefficient of validation sets can be obtained at https://drive.google.com/file/d/13nIzX7YcRdF0-oUrwbAolVEFPEjOI0SE/view?usp=sharing.
After the results are verified, I will use a dataset with 500 training images (400 train, 100 validation) and 200 testing images to get the most accurate model. I also tried using XitoSBML with the inferred images I already had. The images and corresponding SBML documents can be found at https://drive.google.com/drive/folders/1CrGlifucDRInyT-x3sBYo-SmKY1vAQMh?usp=sharing (https://drive.google.com/drive/folders/1CrGlifucDRInyT-x3sBYo-SmKY1vAQMh). The document containing images and the corresponding domain hierarchy can be found at https://drive.google.com/file/d/1wst2dD9xT-tENi_E0dY_wcBWRAY12xbh/view?usp=sharing (https://drive.google.com/file/d/1wst2dD9xT-tENi_E0dY_wcBWRAY12xbh/view).
Until the next time...
Aloha!
Comments
Post a Comment