Community Bonding II
Hi!
Further updates on the work:
1. I have posted the article titled A Beginner's Guide to Spatial Processes package of SBML Version 3 Core 2 on Medium.
Link: https://medium.com/@me1999bh/a-beginners-guide-to-spatial-processes-package-of-sbml-version-3-core-2-2eba4babca11?source=friends_link&sk=cd36b695ab5dd644795bbc279bfd8c62
2. An appropriate dataset of mouse neuroblastoma cells was found and shared by my mentor, Yuta Tokuoka. This dataset consists of wide-field epifluorescent images of cultured neurons with both cytoplasmic (phalloidin) and nuclear stains (DAPI) and a set of manual segmentations of neuronal and nuclear boundaries that can be used as benchmarking data sets for the development of segmentation algorithms.
Featured project: Autosegmentation of cultured neurons
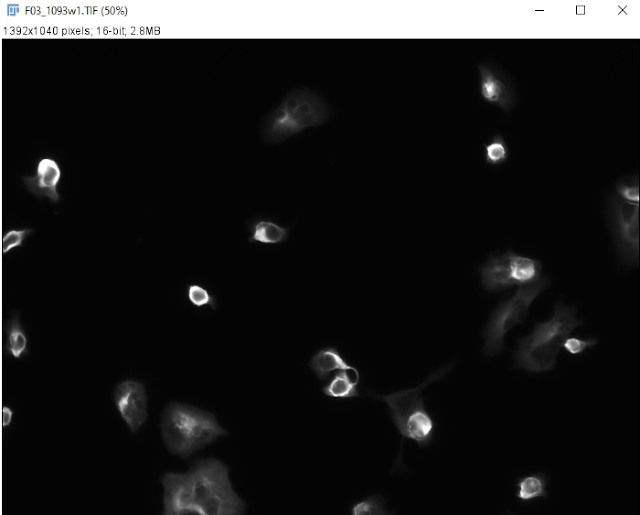 |
| A typical cellular image from the dataset |
 |
| Segmented mask corresponding to the image shown above |
The dataset for this project requires images of single cells along with their segmentation. So, the only work left was to crop individual cells and group them together. So far, I have collected 600 images, each of dimensions 188 X 140 pixels.
 |
| A typical single-cell image and corresponding segmented mask |
3. I was able to build the XitoSBML code and run it on the Eclipse IDE 2020-03R for Java Developers. In this endeavor, I was helped by my mentors Kaito Ii and Akira Funahashi.
As a first step, the source code needs to be cloned.
% git clone https://github.com/spatialsimulator/XitoSBML.git
Further steps are:
1. Import XitoSBML as a Maven project in Eclipse IDE.
2. Right-click the XitoSBML project.
3. Select 'Maven' --> "Update project".
 |
| Running MainImgSpatial.java |
Till the next time
Au revoir!


Comments
Post a Comment